Newsroom
Science Update
Increase in Chromatin Entropy Drives Cellular Aging, Say Researchers
Jun 01, 2022
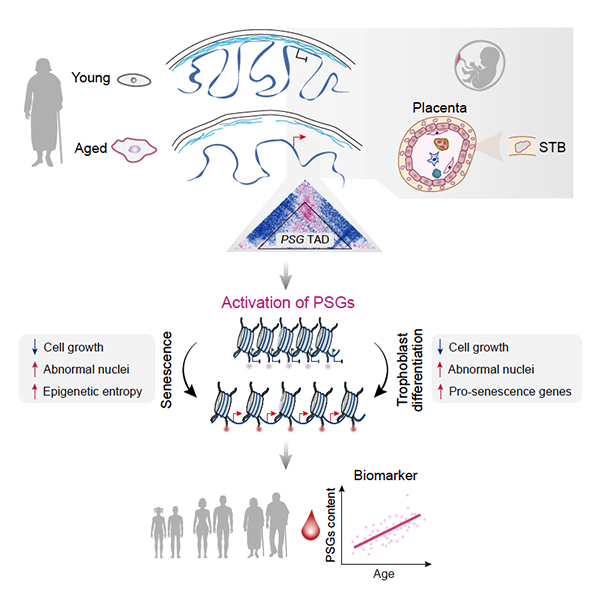
At the center of every cell, highly organized chromatin encodes the program of life with just one set of genes. This is possible because different genes are activated at different stages of life while remaining silent otherwise. Some genes for previous stage of life are buried deep at the nuclear periphery, while some manage to escape from repression. Researchers have been deciphering this hierarchical organization and how it keeps everything in order. However, a new study shows that the system turns chaotic during aging. Specifically, chaos emanates from the nuclear periphery, resulting in large-scale chromatin reorganization and epigenetic dysregulation that ultimately drives cellular senescence. In a new study published on Developmental Cell, researchers from the Institute of Zoology and the Beijing Institute of Genomics of the Chinese Academy of Sciences have collaborated to decode the large-scale remodeling of the epigenomic landscape during aging. They found that aberrant expression of placenta-related genes is the key driver and molecular biomarker of cellular aging. Aging is associated with stem cell exhaustion, the hallmarks of which include a diverse spectrum of epigenetic dysregulation. At the cellular level, aged human stem cells frequently have deformed nuclei. But how this aging phenotype relates to the aged epigenetic state was not well understood. Using isogenic young and senescent, progeroid human mesenchymal progenitor cells (hMPCs) as a cellular aging paradigm, the researchers generated a large-scale, high-resolution epigenomic landscape resource. Through in-depth analyses of reorganization and interplay within and in between hierarchical layers of epigenomic organization during the course of hMPC senescence, the researchers detected large-scale alterations caused by global relaxation of structural constraints in the nuclear lamina and loss of epigenetic markers. They found that the epigenome of senescent cells exhibited increased epigenetic “entropy,” as well as compromised compartmentalization. In addition to clarifying the general relationship between 3D genomic reorganization and transcriptional dysregulation, they identified specific topological conversions that lead to ectopic expression of developmentally restricted genes, e.g., a cluster of pregnancy-specific glycoprotein (PSG) genes. They verified that dissociation of the PSG cluster from the nuclear lamina and consequent reactivation can trigger cellular senescence in aged hMPCs. Moreover, evidence that PSGs serve as novel biomarkers of human organismal aging and are even potential drivers of multiple types of cellular and tissue aging were provided. All in all, the researchers identified the increased epigenetic entropy and activation of developmentally restricted genes as novel hallmarks and driving forces of human cellular aging. These findings provide novel insights into the epigenomic basis of aging regulation and will guide the development of clinical interventions that can slow aging and alleviate aging-associated disorders.
Figure 1. Aberrant activation of developmentally restricted genes caused by large-scale epigenome remodeling is a driver of human stem cell aging (Image from Liu et al. Cell Stem Cell 2022) Original link::https://doi.org/10.1016/j.devcel.2022.05.004 Contact: Guang-Hui Liu Institute of Zoology Chinese Academy of Sciences Tel: 86-64807852 E-mail: ghliu@ioz.ac.cn |
附件: